Benefits
- Compatible with ONT sequencing
- Chimerism levels down to 0.5%
- Results for a single sample in <3 hours
Description
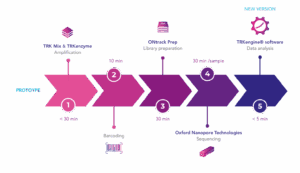
Chimerism monitoring after hematopoietic stem cell transplantation enables prediction of an upcoming relapse. Enhanced sensitivity of chimerism assays allows for earlier detection, thereby increasing the time available for treatment. ONtrack is an in-development solution designed to enable rapid and sensitive chimerism monitoring using Oxford Nanopore Technologies (ONT) platforms. Based on the well-established NGStrack assay, ONtrack uses the same 34 biallelic indel markers and integrates seamlessly into the ONT sequencing workflow. The combination of ONT speed and TRKengine® data analysis enables same-day results for urgent or high-throughput chimerism research.