Benefits
- Regular updates included
- Intuitive interface
- RUO & CE-IVD
Description
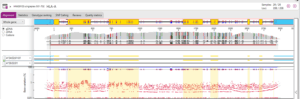
NGSengine® is our robust and intuitive software for high-resolution HLA typing from next-generation sequencing data. It operates efficiently on any standard laptop and automates the complex steps of data analysis, delivering reliable genotyping results. NGSengine® also provides powerful tools to assess data quality, offering both quick overviews and advanced options for detailed inspection.
NGSengine® enables reliable and efficient HLA typing in both research and clinical settings. Available as RUO and CE-marked IVD.
You have questions or want to arrange a demo?
See specifications